Illumina DNA PCR-Free Prep


About the Product
An efficient, fast solution that yields highly accurate data
Our Products
We Provide Various Solutions
- Overview
- Applications
- Documentation
- Compare
- Resources
- Figures
The Illumina DNA PCR-Free Prep workflow supports a broad DNA input range, multiple sample types, and both small and large genomes. The workflow includes DNA extraction from blood, saliva, or dried blood spots.

- Reduces cost by eliminating pre- and post-library quantification steps
- Provides highly uniform coverage across repetitive or uneven genome regions
- Delivers fast, automation-compatible workflow in 90 min
Simplify lab operations
The Illumina DNA PCR-Free Prep workflow supports a broad DNA input range, multiple sample types, and both small and large genomes. The workflow includes DNA extraction from blood, saliva, or dried blood spots.
Obtain reliable results
On-bead tagmentation chemistry combined with PCR-free library preparation eliminates PCR-induced bias and diminishes opportunities for error, providing superior and even coverage across high-GC or -AT regions. Learn more about on-bead tagmentation.
Access flexible throughput
The Illumina DNA/RNA UD Indexes Sets offer up to 384 unique dual indexes, enabling accurate assignment of reads and efficient use of the flow cell.

Prep
- Illumina DNA PCR-Free Prep
Sequence
- MiSeq System
- NextSeq 1000 and 2000
- Systems
- NovaSeq 6000 System
- NovaSeq X Series
Analyze
- DRAGEN secondary analysis
- DRAGEN Germline
- DRAGEN Somatic
- DRAGEN PopGen
- Emedgene
Sequence
- High-performance whole-genome sequencing with Illumina DNA PCR-Free Prep, Tagmentation
- Microbial whole-genome sequencing with Illumina DNA PCR-Free Prep, Tagmentation
- Tunable insert sizes with Illumina DNA PCR-Free Prep, Tagmentation
- Illumina library preparation solutions
- Illumina Genomics Architecture enables PopGen studies with Illumina DNA PCR-Free Prep
- Optimal loading concentrations for Illumina DNA PCR-Free libraries
- Optimal variant calling with Illumina DNA PCR-Free Prep on the NovaSeq X Series
- Illumina DNA PCR-Free Prep, Tagmentation
- Illumina DNA PCR-Free Prep index correction
| Illumina DNA PCR-Free Prep | Illumina DNA Prep | TruSeq DNA PCR-Free | TruSeq DNA Nano |
|---|
Assay time
| ~1.5 hr | ~3-4 hr (from DNA extraction to normalized library) |
|---|
Automation capability
Automation details
| Explore available automation methods | Explore available automation methods | Explore available automation methods | Explore available automation methods |
|---|
Description

This webinar discusses the library preparation workflow and sequencing considerations and provides tips for successful next-generation sequencing projects.
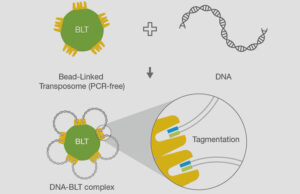
Illumina DNA PCR-Free Prep chemistry
A unique combination of on-bead tagmentation with a PCR-free workflow in a single, rapid reaction.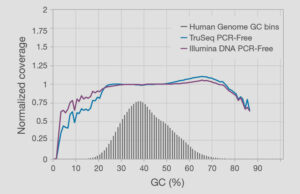
Illumina DNA PCR-Free Prep coverage uniformity
The Illumina DNA PCR-Free Prep provides uniform coverage across a range of GC content in the human genome.
Comparison of read coverage across GC-rich regions
The Illumina DNA PCR-Free Prep kit provides superior read coverage across the GC-rich promoter region of the human RNPEPL1 gene, as compared to TruSeq DNA PCR-Free and TruSeq DNA Nano Library Prep Kits.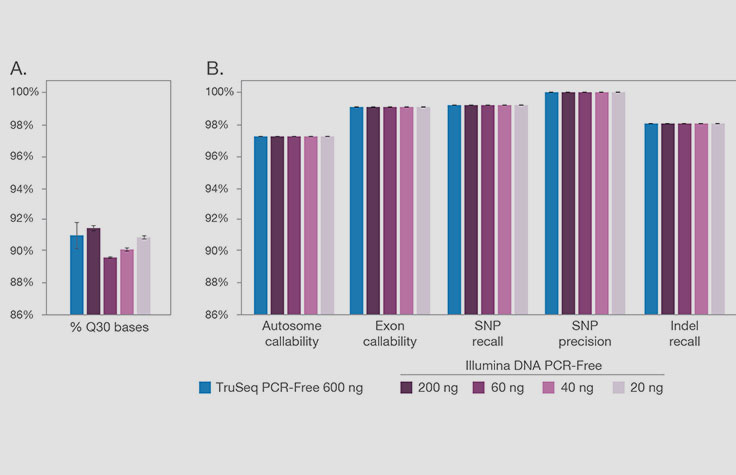
Illumina DNA PCR-Free Prep performance across a range of DNA inputs
Illumina DNA PCR-Free Prep libraries prepared from a range of DNA inputs demonstrate (A) passing quality specifications for all DNA inputs and (B) equivalent callability performance.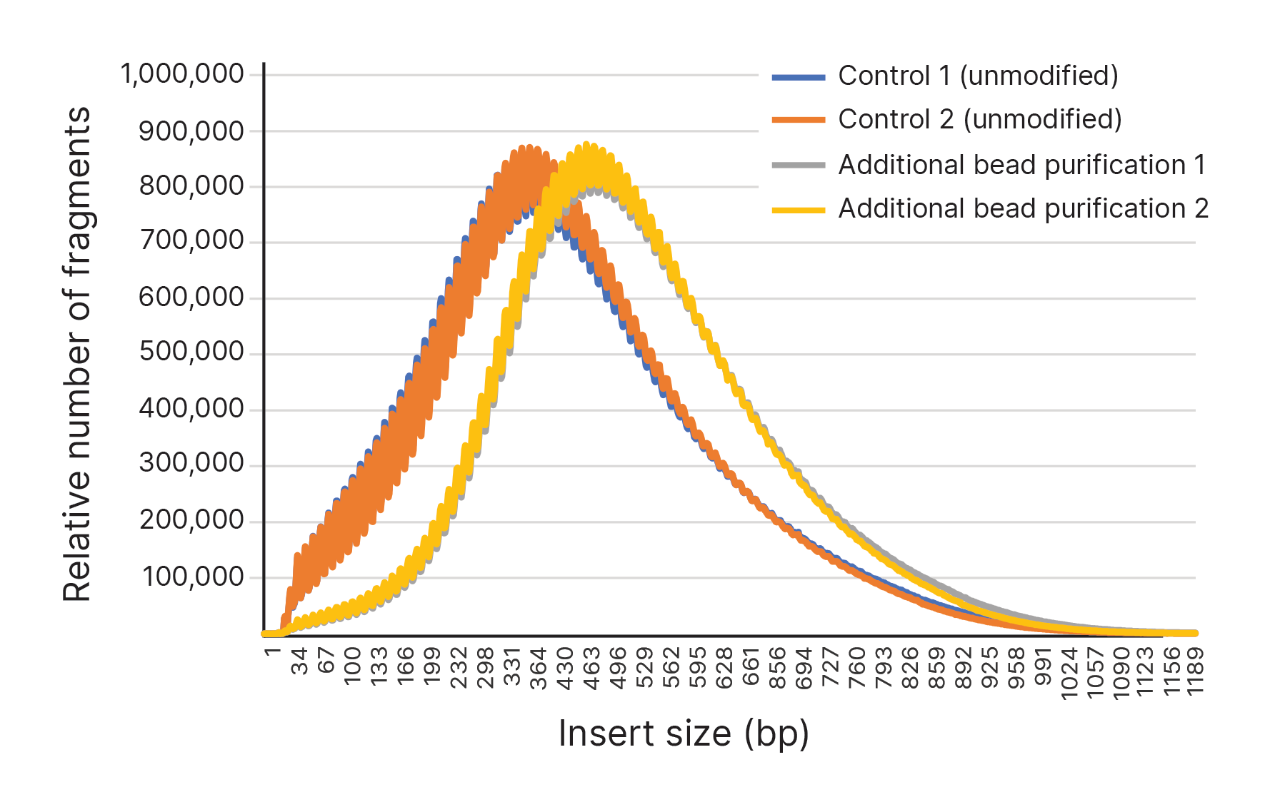
Increased insert size with additional bead purification
An additional round of bead purification results in increased insert sizes (gray and yellow lines), as compared to the unmodified protocol (blue and orange lines).
ELTA 90 is a leading distributor of scientific equipment in South-eastern Europe.
Subscribe Now
Don’t miss our future updates! Get Subscribed Today!

